Doubled barbell
[1]:
%matplotlib inline
[2]:
# Not really required
import sys
sys.path.insert(0, '../../..')
[3]:
from pyLDLE2 import datasets
from pyLDLE2 import buml_, util_
matplotlib.get_backend() = module://matplotlib_inline.backend_inline
[4]:
save_dir_root = '../data/pyLDLE2/doubled_barbell/'
[5]:
X, labelsMat, ddX = datasets.Datasets().barbell()
X.shape = (10227, 2)
[6]:
# The supplied options would override the default options
buml_obj = buml_.BUML(local_opts={'algo':'LDLE', 'N': 200, 'tau': 90},
intermed_opts={'eta_min':5, 'algo':'best'},
vis_opts={'c': labelsMat[:,0], 'save_dir': save_dir_root},
verbose=True, debug=True)
local_opts['k_nn0'] = 700 is created.
Options provided:
local_opts:
{
"Atilde_method": "LDLE_1",
"N": 200,
"U_method": "k_nn",
"algo": "LDLE",
"alpha": 1,
"debug": true,
"delta": 0.9,
"gl_type": "unnorm",
"k": 28,
"k_nn": 49,
"k_nn0": 700,
"k_tune": 7,
"lambda1_decay": 0.75,
"lambda1_init": 8,
"lambda1_min": 0.001,
"max_iter": 300,
"max_sparsity": 0.9,
"metric": "euclidean",
"n_proc": 32,
"p": 0.99,
"power": 5,
"pp_n_thresh": 32,
"radius": 0.5,
"reg": 0.0,
"scale_by": "gamma",
"tau": 90,
"to_postprocess": true,
"verbose": true
}
intermed_opts:
{
"algo": "best",
"debug": true,
"eta_max": 25,
"eta_min": 5,
"len_S_thresh": 256,
"local_algo": "LDLE",
"metric": "euclidean",
"n_proc": 32,
"n_times": 4,
"verbose": true
}
global_opts:
{
"add_dim": false,
"align_transform": "rigid",
"align_w_parent_only": true,
"alpha": 0.3,
"beta": {
"align": null,
"repel": 1
},
"color_tear": true,
"compute_error": false,
"debug": true,
"eps": 1e-08,
"err_tol": 0.0001,
"far_off_points_type": "reuse_fixed",
"init_algo_name": "procrustes",
"k": 28,
"max_internal_iter": 100,
"max_iter": 20,
"metric": "euclidean",
"n_proc": 32,
"n_repel": 0,
"nu": 3,
"patience": 5,
"refine_algo_name": "rgd",
"repel_by": 0.0,
"to_tear": true,
"verbose": true,
"vis_before_init": false
}
[7]:
buml_obj.fit(X=X, ddX=ddX)
Doubling manifold.
No. of points on the boundary = 764
/home/dhruv/anaconda3/lib/python3.9/site-packages/sklearn/neighbors/_base.py:234: EfficiencyWarning: Precomputed sparse input was not sorted by row values. Use the function sklearn.neighbors.sort_graph_by_row_values to sort the input by row values, with warn_when_not_sorted=False to remove this warning.
warnings.warn(
Done.
##############################
Time elapsed from last time log: 41.5 seconds
Total time elapsed: 41.5 seconds
##############################
Computing local views using LDLE
Constructing unnorm graph Laplacian + its eigendecomposition.
Done.
##############################
Time elapsed from last time log: 40.0 seconds
Total time elapsed: 40.0 seconds
##############################
Computing Atilde: Inner Prod of Grad of EigFuncs.
FEM for Atilde.
Atilde: 0 points processed...
Atilde: 4922 points processed...
Atilde: 9844 points processed...
Atilde: 14766 points processed...
Atilde: 19688 points processed...
Atilde: all points processed...
Done.
##############################
Time elapsed from last time log: 27.8 seconds
Total time elapsed: 67.8 seconds
##############################
Computing LDLE.
local_param: all 19690 points processed...
max distortion is 113.043904
Done.
##############################
Time elapsed from last time log: 5.2 seconds
Total time elapsed: 73.4 seconds
##############################
Posprocessing local parameterizations.
#Param replaced: 16656, max distortion: 14.282789
#Param replaced: 3335, max distortion: 14.102668
#Param replaced: 180, max distortion: 14.102668
#Param replaced: 10, max distortion: 14.102668
#Param replaced: 1, max distortion: 14.102668
#Param replaced: 0, max distortion: 14.102668
Max local distortion after postprocessing: 14.102668125100376
Done.
##############################
Time elapsed from last time log: 12.8 seconds
Total time elapsed: 86.2 seconds
##############################
Halving objects.
Done.
##############################
Time elapsed from last time log: 10.1 seconds
Total time elapsed: 101.4 seconds
##############################
Max local distortion = 12.482168183225866
Constructing intermediate views.
eta = 2.
#non-empty views with sz < 2 = 10227
#nodes in views with sz < 2 = 10227
/home/dhruv/codebase/pyLDLE2/docs/ldle_nbks/doubled_manifolds/../../../pyLDLE2/intermed_views_.py:122: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
neigh_ind = np.array(neigh_ind)
Costs computed when eta = 2.
##############################
Time elapsed from last time log: 9.7 seconds
Total time elapsed: 10.5 seconds
##############################
ctr=7712, total_len_S=45601, avg_len_S=5.913
Remaining #nodes in views with sz < 2 = 0
Done with eta = 2.
##############################
Time elapsed from last time log: 179.6 seconds
Total time elapsed: 190.1 seconds
##############################
eta = 3.
#non-empty views with sz < 3 = 737
#nodes in views with sz < 3 = 1474
Costs computed when eta = 3.
##############################
Time elapsed from last time log: 5.7 seconds
Total time elapsed: 195.8 seconds
##############################
ctr=1226, total_len_S=21523, avg_len_S=17.555
Remaining #nodes in views with sz < 3 = 0
Done with eta = 3.
##############################
Time elapsed from last time log: 19.5 seconds
Total time elapsed: 215.4 seconds
##############################
eta = 4.
#non-empty views with sz < 4 = 459
#nodes in views with sz < 4 = 1377
Costs computed when eta = 4.
##############################
Time elapsed from last time log: 5.5 seconds
Total time elapsed: 220.9 seconds
##############################
ctr=1181, total_len_S=29976, avg_len_S=25.382
Remaining #nodes in views with sz < 4 = 0
Done with eta = 4.
##############################
Time elapsed from last time log: 25.9 seconds
Total time elapsed: 246.7 seconds
##############################
eta = 5.
#non-empty views with sz < 5 = 295
#nodes in views with sz < 5 = 1180
Costs computed when eta = 5.
##############################
Time elapsed from last time log: 7.5 seconds
Total time elapsed: 254.3 seconds
##############################
ctr=1031, total_len_S=32074, avg_len_S=31.110
Remaining #nodes in views with sz < 5 = 0
Done with eta = 5.
##############################
Time elapsed from last time log: 34.5 seconds
Total time elapsed: 288.8 seconds
##############################
Pruning and cleaning up.
Done.
##############################
Time elapsed from last time log: 1.7 seconds
Total time elapsed: 290.5 seconds
##############################
After clustering, max distortion is 15.704694
Using rigid transforms for alignment.
Computing laterations scores for overlaps b/w intermed views
Done
##############################
Time elapsed from last time log: 7.1 seconds
Total time elapsed: 7.1 seconds
##############################
Computing a lateration.
Seq of intermediate views and their predecessors computed.
No. of connected components = 1
Done.
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 7.1 seconds
##############################
Computing initial embedding using: procrustes algorithm
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 7.1 seconds
##############################
Initial alignment of 316 views completed
Initial alignment of 632 views completed
Initial alignment of 948 views completed
Initial alignment of 1264 views completed
Embedding initialized.
##############################
Time elapsed from last time log: 1.9 seconds
Total time elapsed: 9.0 seconds
##############################
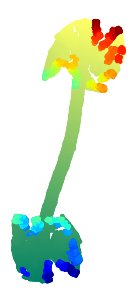
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 0
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 16.9 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 15.5 seconds
Total time elapsed: 32.4 seconds
##############################
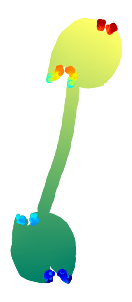
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 1
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 36.6 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 14.2 seconds
Total time elapsed: 50.8 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 2
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 53.4 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 14.7 seconds
Total time elapsed: 68.1 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 3
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 69.6 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 13.6 seconds
Total time elapsed: 83.1 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 4
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 84.9 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 13.5 seconds
Total time elapsed: 98.4 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 5
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 99.9 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 15.6 seconds
Total time elapsed: 115.5 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 6
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 117.1 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 13.3 seconds
Total time elapsed: 130.4 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 7
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 132.5 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 13.2 seconds
Total time elapsed: 145.7 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 8
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 147.2 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 16.7 seconds
Total time elapsed: 163.9 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 9
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 165.5 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 12.7 seconds
Total time elapsed: 178.3 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 10
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 179.7 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 13.1 seconds
Total time elapsed: 192.8 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 11
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 194.3 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 15.3 seconds
Total time elapsed: 209.7 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 12
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 211.9 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 13.1 seconds
Total time elapsed: 225.0 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 13
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 226.3 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 13.5 seconds
Total time elapsed: 239.9 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 14
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 241.2 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 11.5 seconds
Total time elapsed: 252.7 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 15
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 254.1 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 11.6 seconds
Total time elapsed: 265.7 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 16
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 266.9 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 11.3 seconds
Total time elapsed: 278.1 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 17
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 279.5 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 11.1 seconds
Total time elapsed: 290.5 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 18
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 292.0 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 11.5 seconds
Total time elapsed: 303.5 seconds
##############################
Refining with rgd algorithm for 100 iterations.
Refinement iteration: 19
##############################
Time elapsed from last time log: 0.0 seconds
Total time elapsed: 304.7 seconds
##############################
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Descent starts
Done.
##############################
Time elapsed from last time log: 11.2 seconds
Total time elapsed: 315.9 seconds
##############################
Computing error.
min and max weights: 1.0 1.0
Computing Pseudoinverse of a matrix of L of size 10227 + 1266 multiplied with B
Computing svd
Done
Alignment error: 1.130367
##############################
Time elapsed from last time log: 4.5 seconds
Total time elapsed: 320.5 seconds
##############################
[7]:
array([[-0.20815434, -1.38056597],
[-0.19768424, -1.38399708],
[-0.19105738, -1.38372242],
...,
[ 0.08417931, 0.70909419],
[ 0.09127498, 0.70620604],
[ 0.09849001, 0.70241038]])
[8]:
buml_obj.GlobalViews.vis_embedding(buml_obj.GlobalViews.y_final, buml_obj.vis, buml_obj.vis_opts,
buml_obj.GlobalViews.color_of_pts_on_tear_final,
title='Final global embedding', )
[9]:
util_.save(save_dir_root, 'ldle.dat', [X, labelsMat, buml_obj])
[ ]: